
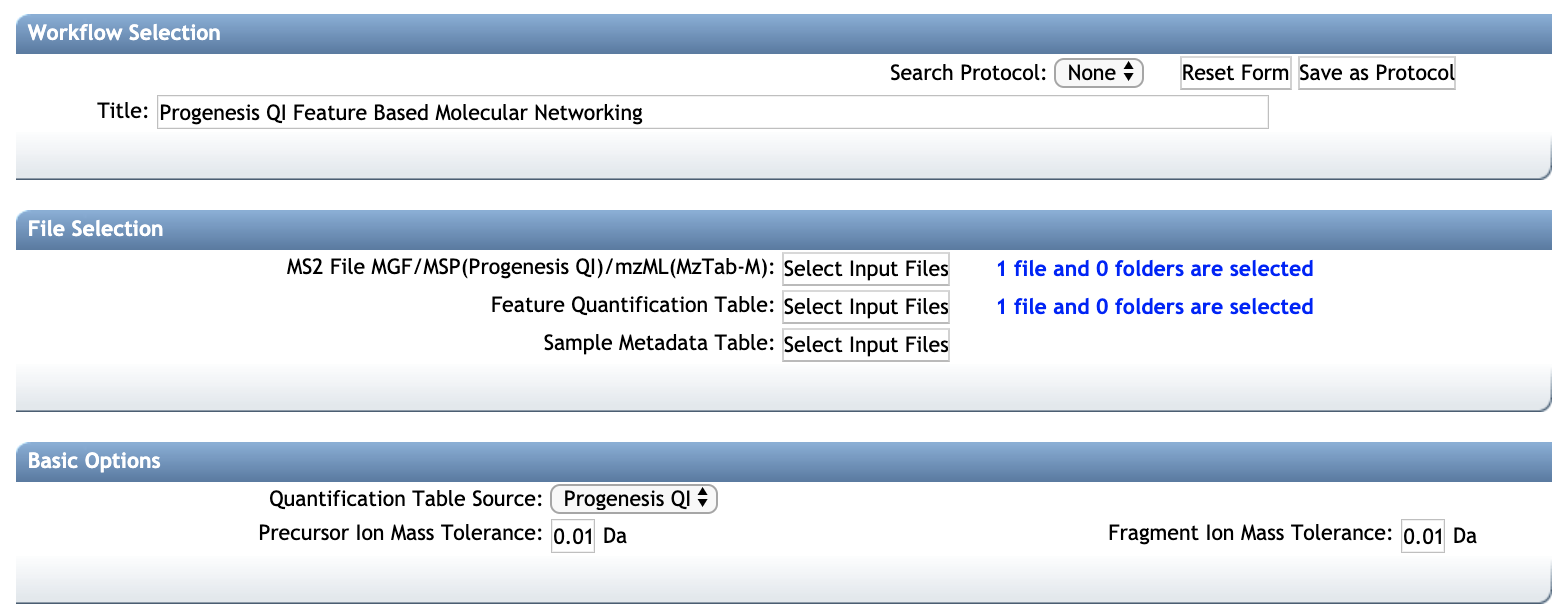
Select your Species and Confidence (score) cutoff and enter the genes or proteins of interest in the Enter. The dialog shown in Figure 1 should then appear. and selecting String DB for the Data Source.
#Cytoscape import csv free#
Into this (feel free to scroll past) The output isn’t too interesting…yet. After installing stringApp from the Cytoscape app store STRING networks may be imported by going to FileImportNetworkPublic databases. Use the drop-down next to the column header to define group1 as a source node (green dot) and group2 as a target node (orange circle). Now we need to add some semantics to the columns in the file. In the following dialog open group-edges.csv. Use data source plugins when you want to import data from. It was a quick and dirty creation to produce a quick result. By adding a data source plugin, you can immediately use the data in any of your existing dashboards. So I began by exporting my pcap data to a csv. Wireshark is a great tool, but as the size of the packet capture increases, the performance drops as it tries to parse the potential hundreds of thousands of lines of traffic data. This example will focus on network packet captures, but a bit of Python applied in the right doses could adapt this project to any network mapping command line tool output. Various tools can be used for this, netflow, nmap, tracert. Importing timeSeries from csv file into R with correct dates. So we try to create the ‘ground truth’ of what the network actually looks like. Solved-Convert Adjacency Matrix into Edgelist (csv file) for Cytoscape-R.
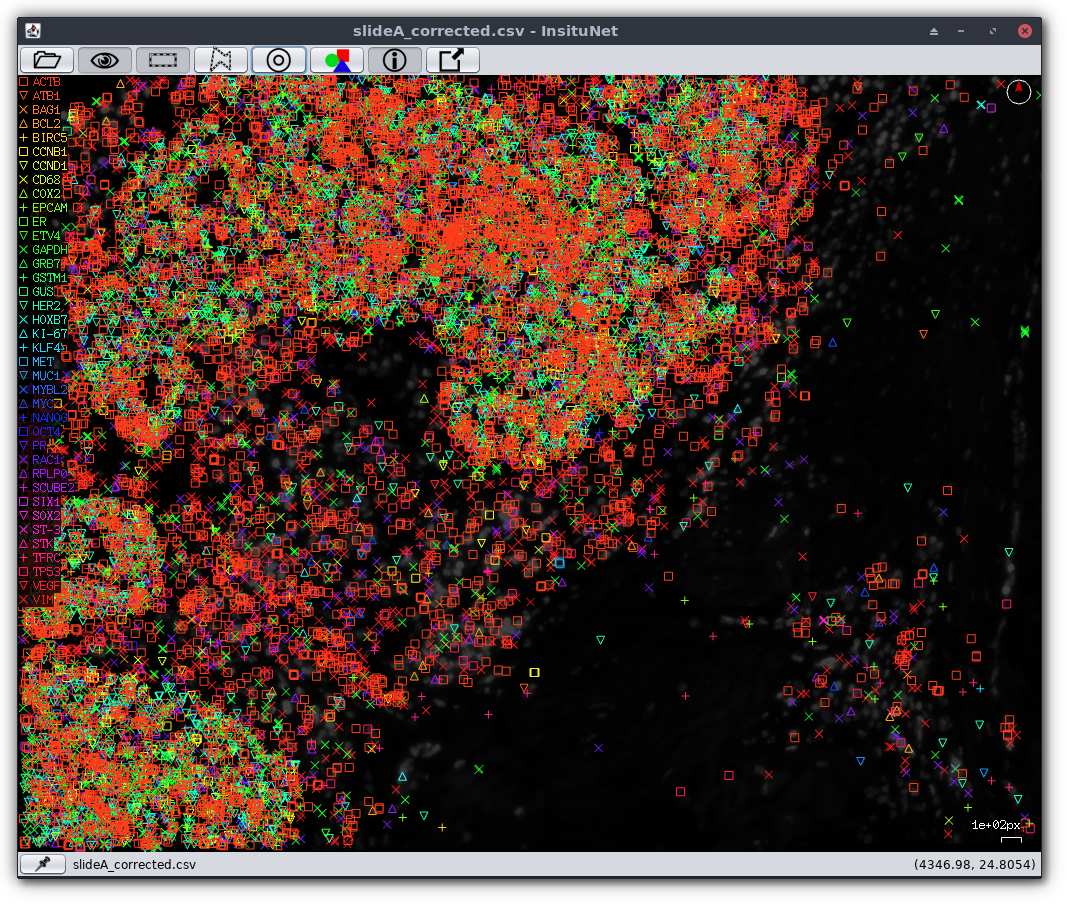
Too often an Incident Responder asks for a map of the network, only to discover it’s more than 2 years old and not much more than a ghost of the current network topology.


 0 kommentar(er)
0 kommentar(er)
